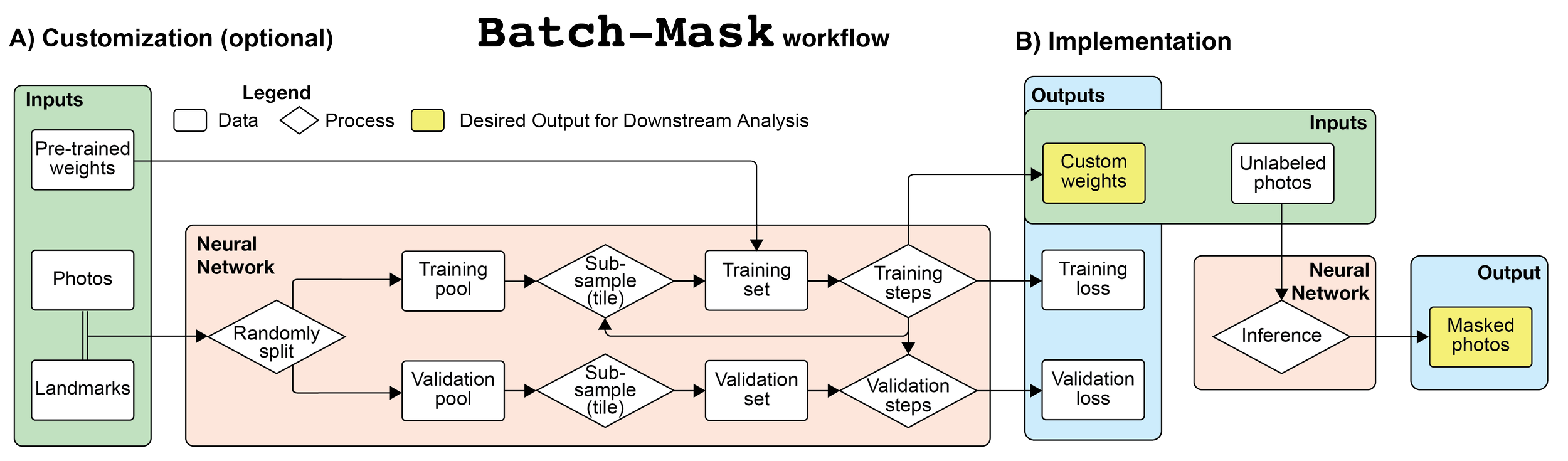
Batch-Mask: An automated workflow using Mask R-CNN to isolate non-standard biological specimens for color analysis
Efficient comparisons of biological color patterns are critical for understanding the mechanisms by which organisms evolve in ecosystems, including sexual selection, predator-prey interactions, and thermoregulation. However, limbless, elongate, or spiral-shaped organisms do not conform to the standard orientation and photographic techniques required for many automated analyses. Currently, large-scale color analysis of elongate animals requires time-consuming manual landmarking, which reduces their representation in coloration research despite their ecological importance. We present Batch-Mask: an automated, customizable workflow to automatically analyze large photographic datasets to isolate non-standard biological organisms from the background. Batch-Mask is completely open-source and does not depend on any proprietary software. We also present a user guide for fine-tuning weights to a custom dataset and incorporating existing manual visual analysis tools (e.g., Mica Toolbox) into a single automated workflow for comparing color patterns across images. Batch-Mask was 60x faster than manual landmarking and produced masks that correctly identified 96% of all snake pixels. To validate our approach, we used the micatoolbox to compare pattern energy in a sample set of snake photographs segmented by Batch-Mask and humans and found no significant difference in the output results. The fine-tuned weights, user guide, and automated workflow substantially decrease the amount of time and attention required to quantitatively analyze non-standard biological subjects. With these tools, biologists can compare color, pattern, and shape differences in large datasets that include significant morphological variation in elongate body forms.This advance is especially valuable for comparative analyses of natural history collections across a broad range of morphologies. Through automation, Batch-Mask can greatly expand the scale of space, time, or taxonomic breadth across which color variation can be quantitatively examined.
@article{CurlisBatchMask, author = {{John David} Curlis and Timothy Renney and Alison R. {Davis Rabosky} and Talia Yuki Moore}, year = {2022}, journal = {Integrative and Comparative Biology}, title = {{Batch-Mask: An automated workflow using Mask R-CNN to isolate non-standard biological specimens for color analysis}}}